The Original Mouse Models of Glioblastoma: Analysis of Pathophysiological Characteristics of Transplanted Tumor Tissue
The aim of this study was to morphologically, molecularly, and immunologically characterize two new transplantable glioblastoma (GB) tissue models, designated M2 GB and M6 GB.
Materials and Methods. Two new chemically induced, easily transplantable tissue mouse models of high-grade glioma have been created and characterized. M2 GB and M6 GB tissues were orthotopically transplanted to immunocompetent C57BL/6 mice. The clinical and morphological characteristics of tumor growth, as well as the intratumoral immune response and target gene expression were assessed.
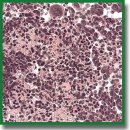
Results. Clinical manifestations of M2 GB and M6 GB growth in mice include motility disorders, cachexia, and priapism. Morphologically, M2 GB and M6 GB are characterized by diffuse proliferation, cellular and nuclear polymorphism, and high mitotic activity with pathological mitotic patterns corresponding to the aggressive nature of the mentioned tumors. Both tumors were significantly infiltrated with CD3+ T lymphocytes (~32%) and F4/80+ macrophages (~28–50%). M2 GB showed a higher content of F4/80+ macrophages compared to M6 GB. The Cdkn2a, S100b, Mki67, Pten, Vegfa, Hif1a, Sox2, Abcb1, and Gfap genes were overexpressed in both tumors. Expression of the Cd133, Tp53, and Pdgfra genes was increased in M2 GB. High expression of Pi3k and Gdnf was seen in M6 GB. Expression of Cd44, Pi3k, Hif1a, Gdnf, and Egfr was higher in M6 GB tissues compared to M2 GB, whereas expression of Cdkn2a, Tp53, Cd133,and Pdgfra was higher in M2 GB tissues compared to M6 GB.
Conclusion. The M2 GB and M6 GB models of transplanted tissues reproduce key characteristics of human GB, including similar intracellular immune profiles, clinical and morphological features, and gene expression patterns, which are important for further research in neurological oncology. These models can be used to develop diagnostic and treatment methods and to study tumor genesis.
- Wu W., Klockow J.L., Zhang M., Lafortune F., Chang E., Jin L., Wu Y., Daldrup-Link H.E. Glioblastoma multiforme (GBM): an overview of current therapies and mechanisms of resistance. Pharmacol Res 2021; 171: 105780, https://doi.org/10.1016/j.phrs.2021.105780.
- Background lesions in laboratory animals. A color atlas. McInnes E.F., Mann P. (editors). Elsevier Ltd.; 2011, https://doi.org/10.1016/C2009-0-41283-2.
- Lampreht Tratar U., Horvat S., Cemazar M. Transgenic mouse models in cancer research. Front Oncol 2018; 8: 268, https://doi.org/10.3389/fonc.2018.00268.
- Ireson C.R., Alavijeh M.S., Palmer A.M., Fowler E.R., Jones H.J. The role of mouse tumour models in the discovery and development of anticancer drugs. Br J Cancer 2019; 121(2): 101–108, https://doi.org/10.1038/s41416-019-0495-5.
- Sahu U., Barth R.F., Otani Y., McCormack R., Kaur B. Rat and mouse brain tumor models for experimental neuro-oncology research. J Neuropathol Exp Neurol 2022; 81(5): 312–329, https://doi.org/10.1093/jnen/nlac021.
- Haddad A.F., Young J.S., Amara D., Berger M.S., Raleigh D.R., Aghi M.K., Butowski N.A. Mouse models of glioblastoma for the evaluation of novel therapeutic strategies. Neurooncol Adv 2021; 3(1): vdab100, https://doi.org/10.1093/noajnl/vdab100.
- Lowenstein P.R., Castro M.G. Uncertainty in the translation of preclinical experiments to clinical trials. Why do most phase III clinical trials fail? Curr Gene Ther 2009; 9(5): 368–374, https://doi.org/10.2174/156652309789753392.
- Fogel D.B. Factors associated with clinical trials that fail and opportunities for improving the likelihood of success: a review. Contemp Clin Trials Commun 2018; 11: 156–164, https://doi.org/10.1016/j.conctc.2018.08.001.
- Mirzayans R., Murray D. What are the reasons for continuing failures in cancer therapy? Are misleading/inappropriate preclinical assays to be blamed? Might some modern therapies cause more harm than benefit? Int J Mol Sci 2022; 23(21): 13217, https://doi.org/10.3390/ijms232113217.
- Arutyunyan I.V., Soboleva A.G., Kovtunov E.A., Kosyreva A.M., Kudelkina V.V., Alekseeva A.I., Elchaninov A.V., Jumaniyazova E.D., Goldshtein D.V., Bolshakova G.B., Fatkhudinov T.K. Gene expression profile of 3D spheroids in comparison with 2D cell cultures and tissue strains of diffuse high-grade gliomas. Bull Exp Biol Med 2023; 175(4): 576–584, https://doi.org/10.1007/s10517-023-05906-y.
- Alekseeva A., Drozd S., Nikitin P., Postnov A., Lipengolts A., Skribitsky V., Finogenova Y., Shpakova K., Khalansky A., Pronin I., Pavlova G. Comparative morphological and molecular genetic characteristics of experimental glioblastoma 101/8 and C6. In: Neuroscience for medicine and psychology. Sudak; 2023; p. 34–35, https://doi.org/10.29003/m3154.sudak.ns2023-19/34-35.
- Paster E.V., Villines K.A., Hickman D.L. Endpoints for mouse abdominal tumor models: refinement of current criteria. Comp Med 2009; 59(3): 234–241.
- Fedoseeva V.V., Khalansky A.S., Mkhitarov V.A., Tsvetkov I.S., Malinovskaya Y.A., Maksimenko O.O., Gelperina S.E., Balabanyan V.Y., Razzhivina V.A., Gorelikov P.L., Mikhailova L.P., Makarova O.V. Anti-tumor activity of doxorubicin-loaded poly(lactide-co-glycolide) nanoparticles in the experimental glioblastoma. Klinicheskaya i eksperimental’naya morfologiya 2017; 2(22): 65–71.
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29(9): e45, https://doi.org/10.1093/nar/29.9.e45.
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3(7): RESEARCH0034, https://doi.org/10.1186/gb-2002-3-7-research0034.
- Inoue A., Ohnishi T., Nishikawa M., Watanabe H., Kusakabe K., Taniwaki M., Yano H., Ohtsuka Y., Matsumoto S., Suehiro S., Yamashita D., Shigekawa S., Takahashi H., Kitazawa R., Tanaka J., Kunieda T. Identification of CD44 as a reliable biomarker for glioblastoma invasion: based on magnetic resonance imaging and spectroscopic analysis of 5-aminolevulinic acid fluorescence. Biomedicines 2023; 11(9): 2369, https://doi.org/10.3390/biomedicines11092369.
- Wang L.J., Lv P., Lou Y. Alarm signal S100-related signature is correlated with tumor microenvironment and predicts prognosis in glioma. Dis Markers 2022; 2022: 4968555, https://doi.org/10.1155/2022/4968555.
- Yuan Q., Zuo F.X., Cai H.Q., Qian H.P., Wan J.H. Identifying differential expression genes and prognostic signature based on subventricular zone involved glioblastoma. Front Genet 2022; 13: 912227, https://doi.org/10.3389/fgene.2022.912227.
- Luo X., Xu S., Zhong Y., Tu T., Xu Y., Li X., Wang B., Yang F. High gene expression levels of VEGFA and CXCL8 in the peritumoral brain zone are associated with the recurrence of glioblastoma: a bioinformatics analysis. Oncol Lett 2019; 18(6): 6171–6179, https://doi.org/10.3892/ol.2019.10988.
- Liu W., Lv G., Li Y., Li L., Wang B. Downregulation of CDKN2A and suppression of cyclin D1 gene expressions in malignant gliomas. J Exp Clin Cancer Res 2011; 30(1): 76, https://doi.org/10.1186/1756-9966-30-76.
- Shiraishi S., Tada K., Nakamura H., Makino K., Kochi M., Saya H., Kuratsu J., Ushio Y. Influence of p53 mutations on prognosis of patients with glioblastoma. Cancer 2002; 95(2): 249–257, https://doi.org/10.1002/cncr.10677.
- Dahlrot R.H., Bangsø J.A., Petersen J.K., Rosager A.M., Sørensen M.D., Reifenberger G., Hansen S., Kristensen B.W. Prognostic role of Ki-67 in glioblastomas excluding contribution from non-neoplastic cells. Sci Rep 2021; 11(1): 17918, https://doi.org/10.1038/s41598-021-95958-9.
- Hashemi M., Etemad S., Rezaei S., Ziaolhagh S., Rajabi R., Rahmanian P., Abdi S., Koohpar Z.K., Rafiei R., Raei B., Ahmadi F., Salimimoghadam S., Aref A.R., Zandieh M.A., Entezari M., Taheriazam A., Hushmandi K. Progress in targeting PTEN/PI3K/Akt axis in glioblastoma therapy: revisiting molecular interactions. Biomed Pharmacother 2023; 158: 114204, https://doi.org/10.1016/j.biopha.2022.114204.
- Sfifou F., Hakkou E.M., Bouaiti E.A., Slaoui M., Errihani H., Al Bouzidi A., Abouqal R., El Ouahabi A., Cherradi N. Correlation of immunohistochemical expression of HIF-1alpha and IDH1 with clinicopathological and therapeutic data of moroccan glioblastoma and survival analysis. Ann Med Surg (Lond) 2021; 69: 102731, https://doi.org/10.1016/j.amsu.2021.102731.
- Yu W., Ren X., Hu C., Tan Y., Shui Y., Chen Z., Zhang L., Peng J., Wei Q. Glioma SOX2 expression decreased after adjuvant therapy. BMC Cancer 2019; 19(1): 1087, https://doi.org/10.1186/s12885-019-6292-y.
- Abdoli Shadbad M., Nejadi Orang F., Baradaran B. CD133 significance in glioblastoma development: in silico and in vitro study. Eur J Med Res 2024; 29(1): 154, https://doi.org/10.1186/s40001-024-01754-2.
- Farsi Z., Allahyari Fard N. The identification of key genes and pathways in glioblastoma by bioinformatics analysis. Mol Cell Oncol 2023; 10(1): 2246657, https://doi.org/10.1080/23723556.2023.2246657.
- Yu Z., Li H., Wang M., Luo W., Xue Y. GDNF regulates lipid metabolism and glioma growth through RET/ERK/HIF-1/SREBP-1. Int J Oncol 2022; 61(3): 109, https://doi.org/10.3892/ijo.2022.5399.
- Szylberg M., Sokal P., Śledzińska P., Bebyn M., Krajewski S., Szylberg Ł., Szylberg A., Szylberg T., Krystkiewicz K., Birski M., Harat M., Włodarski R., Furtak J. MGMT promoter methylation as a prognostic factor in primary glioblastoma: a single-institution observational study. Biomedicines 2022, https://doi.org/10.3390/biomedicines10082030.
- Roy L.O., Lemelin M., Blanchette M., Poirier M.B., Aldakhil S., Fortin D. Expression of ABCB1, ABCC1 and 3 and ABCG2 in glioblastoma and their relevance in relation to clinical survival surrogates. J Neurooncol 2022; 160(3): 601–609, https://doi.org/10.1007/s11060-022-04179-1.
- Ahmadipour Y., Gembruch O., Pierscianek D., Sure U., Jabbarli R. Does the expression of glial fibrillary acid protein (GFAP) stain in glioblastoma tissue have a prognostic impact on survival? Neurochirurgie 2020; 66(3): 150–154, https://doi.org/10.1016/j.neuchi.2019.12.012.
- Dhawan A., Manem V.S.K., Yeaney G., Lathia J.D., Ahluwalia M.S. EGFR pathway expression persists in recurrent glioblastoma independent of amplification status. Cancers (Basel) 2023; 15(3): 670, https://doi.org/10.3390/cancers15030670.
- Goldman O., Adler L.N., Hajaj E., Croese T., Darzi N., Galai S., Tishler H., Ariav Y., Lavie D., Fellus-Alyagor L., Oren R., Kuznetsov Y., David E., Jaschek R., Stossel C., Singer O., Malitsky S., Barak R., Seger R., Erez N., Amit I., Tanay A., Saada A., Golan T., Rubinek T., Sang Lee J., Ben-Shachar S., Wolf I., Erez A. Early Infiltration of innate immune cells to the liver depletes HNF4α and promotes extrahepatic carcinogenesis. Cancer Discov 2023; 13(7): 1616–1635, https://doi.org/10.1158/2159-8290.CD-22-1062.
- Law M.L. Cancer cachexia: pathophysiology and association with cancer-related pain. Front Pain Res (Lausanne) 2022; 3: 971295, https://doi.org/10.3389/fpain.2022.971295.
- Olson B., Diba P., Korzun T., Marks D.L. Neural mechanisms of cancer cachexia. Cancers (Basel) 2021; 13(16): 3990, https://doi.org/10.3390/cancers13163990.
- Zhong W., Jina H., Rathore P., Wong E.L., Mancuso P., Lalak N., Hayden L., Haghighi K. A case report of priapism with unusual presentation and clinical course. Urol Case Rep 2017; 12: 70–72, https://doi.org/10.1016/j.eucr.2017.03.009.
- Shelton L.M., Mukherjee P., Huysentruyt L.C., Urits I., Rosenberg J.A., Seyfried T.N. A novel pre-clinical in vivo mouse model for malignant brain tumor growth and invasion. J Neurooncol 2010; 99(2): 165–176, https://doi.org/10.1007/s11060-010-0115-y.
- Cui P., Shao W., Huang C., Wu C.J., Jiang B., Lin D. Metabolic derangements of skeletal muscle from a murine model of glioma cachexia. Skelet Muscle 2019; 9(1): 3, https://doi.org/10.1186/s13395-018-0188-4.
- Monteiro R.Q., Lima L.G., Gonçalves N.P., De Souza M.R., Leal A.C., Demasi M.A., Sogayar M.C., Carneiro-Lobo T.C. Hypoxia regulates the expression of tissue factor pathway signaling elements in a rat glioma model. Oncol Lett 2016; 12(1): 315–322, https://doi.org/10.3892/ol.2016.4593.
- Onizuka H., Masui K., Komori T. Diffuse gliomas to date and beyond 2016 WHO Classification of Tumours of the central nervous system. Int J Clin Oncol 2020; 25(6): 997–1003, https://doi.org/10.1007/s10147-020-01695-w.
- Himes B.T., Geiger P.A., Ayasoufi K., Bhargav A.G., Brown D.A., Parney I.F. Immunosuppression in glioblastoma: current understanding and therapeutic implications. Front Oncol 2021; 11: 770561, https://doi.org/10.3389/fonc.2021.770561.
- Alghamri M.S., McClellan B.L., Hartlage C.S., Haase S., Faisal S.M., Thalla R., Dabaja A., Banerjee K., Carney S.V., Mujeeb A.A., Olin M.R., Moon J.J., Schwendeman A., Lowenstein P.R., Castro M.G. Targeting neuroinflammation in brain cancer: uncovering mechanisms, pharmacological targets, and neuropharmaceutical developments. Front Pharmacol 2021; 12: 680021, https://doi.org/10.3389/fphar.2021.680021.
- Andersen J.K., Miletic H., Hossain J.A. Tumor-associated macrophages in gliomas-basic insights and treatment opportunities. Cancers (Basel) 2022; 14(5): 1319, https://doi.org/10.3390/cancers14051319.
- Georgieva P.B., Mathivet T., Alt S., Giese W., Riva M., Balcer M., Gerhardt H. Long-lived tumor-associated macrophages in glioma. Neurooncol Adv 2020; 2(1): vdaa127, https://doi.org/10.1093/noajnl/vdaa127.
- Rodrigues L.F., Camacho A.H.D.S., Spohr T.C.L.S.E. Secondary glioblastoma metastasis outside the central nervous system in a young HIV-infected patient. Ther Adv Med Oncol 2020; 12: 1758835920923432, https://doi.org/10.1177/1758835920923432.
- Chongsathidkiet P., Jackson C., Koyama S., Loebel F., Cui X., Farber S.H., Woroniecka K., Elsamadicy A.A., Dechant C.A., Kemeny H.R., Sanchez-Perez L., Cheema T.A., Souders N.C., Herndon J.E., Coumans J.V., Everitt J.I., Nahed B.V., Sampson J.H., Gunn M.D., Martuza R.L., Dranoff G., Curry W.T., Fecci P.E. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med 2018; 24(9): 1459–1468, https://doi.org/10.1038/s41591-018-0135-2.
- Sanchez V.E., Lynes J.P., Walbridge S., Wang X., Edwards N.A., Nwankwo A.K., Sur H.P., Dominah G.A., Obungu A., Adamstein N., Dagur P.K., Maric D., Munasinghe J., Heiss J.D., Nduom E.K. GL261 luciferase-expressing cells elicit an anti-tumor immune response: an evaluation of murine glioma models. Sci Rep 2020; 10(1): 11003, https://doi.org/10.1038/s41598-020-67411-w.
- Brodin P., Davis M.M. Human immune system variation. Nat Rev Immunol 2017; 17(1): 21–29, https://doi.org/10.1038/nri.2016.125.
- Maire C.L., Mohme M., Bockmayr M., Fita K.D., Riecken K., Börnigen D., Alawi M., Failla A., Kolbe K., Zapf S., Holz M., Neumann K., Dührsen L., Lange T., Fehse B., Westphal M., Lamszus K. Glioma escape signature and clonal development under immune pressure. J Clin Invest 2020; 130(10): 5257–5271, https://doi.org/10.1172/JCI138760.
- Brown C.E., Hibbard J.C., Alizadeh D., Blanchard M.S., Natri H.M., Wang D., Ostberg J.R., Aguilar B., Wagner J.R., Paul J.A., Starr R., Wong R.A., Chen W., Shulkin N., Aftabizadeh M., Filippov A., Chaudhry A., Ressler J.A., Kilpatrick J., Myers-McNamara P., Chen M., Wang L.D., Rockne R.C., Georges J., Portnow J., Barish M.E., D’Apuzzo M., Banovich N.E., Forman S.J., Badie B. Locoregional delivery of IL-13Rα2-targeting CAR-T cells in recurrent high-grade glioma: a phase 1 trial. Nat Med 2024; 30(4): 1001–1012, https://doi.org/10.1038/s41591-024-02875-1.
- Sobhani N., Bouchè V., Aldegheri G., Rocca A., D’Angelo A., Giudici F., Bottin C., Donofrio C.A., Pinamonti M., Ferrari B., Panni S., Cominetti M., Aliaga J., Ungari M., Fioravanti A., Zanconati F., Generali D. Analysis of PD-L1 and CD3 expression in glioblastoma patients and correlation with outcome: a single center report. Biomedicines 2023; 11(2): 311, https://doi.org/10.3390/biomedicines11020311.
- Musca B., Russo M.G., Tushe A., Magri S., Battaggia G., Pinton L., Bonaudo C., Della Puppa A., Mandruzzato S. The immune cell landscape of glioblastoma patients highlights a myeloid-enriched and immune suppressed microenvironment compared to metastatic brain tumors. Front Immunol 2023; 14: 1236824, https://doi.org/10.3389/fimmu.2023.1236824.
- Nasti T.H., Cochran J.B., Tsuruta Y., Yusuf N., McKay K.M., Athar M., Timares L., Elmets C.A. A murine model for the development of melanocytic nevi and their progression to melanoma. Mol Carcinog 2016; 55(5): 646–658, https://doi.org/10.1002/mc.22310.
- Todorova V.K., Kaufmann Y., Luo S., Klimberg V.S. Modulation of p53 and c-myc in DMBA-induced mammary tumors by oral glutamine. Nutr Cancer 2006; 54(2): 263–273, https://doi.org/10.1207/s15327914nc5402_13.
- Fidianingsih I., Aryandono T., Widyarini S., Herwiyanti S. Profile of histopathological type and molecular subtypes of mammary cancer of DMBA-induced rat and its relevancy to human breast cancer. J Med Sci 2022; 10(A): 71–78, https://doi.org/10.3889/oamjms.2022.7975.
- Robbins D., Wittwer J.A., Codarin S., Circu M.L., Aw T.Y., Huang T.T., Van Remmen H., Richardson A., Wang D.B., Witt S.N., Klein R.L., Zhao Y. Isocitrate dehydrogenase 1 is downregulated during early skin tumorigenesis which can be inhibited by overexpression of manganese superoxide dismutase. Cancer Sci 2012; 103(8): 1429–1433, https://doi.org/10.1111/j.1349-7006.2012.02317.x.
- Evidence based practice in neuro-oncology. Mallick S., Giridhar P., Rath G.K. (editors). Springer Singapore; 2021, https://doi.org/10.1007/978-981-16-2659-3.
- Marker D.F., Agnihotri S., Amankulor N., Murdoch G.H., Pearce T.M. The dominant TP53 hotspot mutation in IDH -mutant astrocytoma, R273C, has distinctive pathologic features and sex-specific prognostic implications. Neurooncol Adv 2021; 4(1): vdab182, https://doi.org/10.1093/noajnl/vdab182.
- Butler M., Pongor L., Su Y.T., Xi L., Raffeld M., Quezado M., Trepel J., Aldape K., Pommier Y., Wu J. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 2020; 6(5): 380–391, https://doi.org/10.1016/j.trecan.2020.02.010.
- Ohtsuki S., Kamoi M., Watanabe Y., Suzuki H., Hori S., Terasaki T. Correlation of induction of ATP binding cassette transporter A5 (ABCA5) and ABCB1 mRNAs with differentiation state of human colon tumor. Biol Pharm Bull 2007; 30(6): 1144–1146, https://doi.org/10.1248/bpb.30.1144.
- Oda Y., Saito T., Tateishi N., Ohishi Y., Tamiya S., Yamamoto H., Yokoyama R., Uchiumi T., Iwamoto Y., Kuwano M., Tsuneyoshi M. ATP-binding cassette superfamily transporter gene expression in human soft tissue sarcomas. Int J Cancer 2005; 114(6): 854–862, https://doi.org/10.1002/ijc.20589.
- Ahmed S., Khan H., Aschner M., Mirzae H., Küpeli Akkol E., Capasso R. Anticancer potential of furanocoumarins: mechanistic and therapeutic aspects. Int J Mol Sci 2020; 21(16): 5622, https://doi.org/10.3390/ijms21165622.
- Robey R.W., Massey P.R., Amiri-Kordestani L., Bates S.E. ABC transporters: unvalidated therapeutic targets in cancer and the CNS. Anticancer Agents Med Chem 2010; 10(8): 625–633, https://doi.org/10.2174/187152010794473957.
- Amiri-Kordestani L., Basseville A., Kurdziel K., Fojo A.T., Bates S.E. Targeting MDR in breast and lung cancer: discriminating its potential importance from the failure of drug resistance reversal studies. Drug Resist Updat 2012; 15(1-2): 50–61, https://doi.org/10.1016/j.drup.2012.02.002.
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Pontén F. Tissue-based map of the human proteome. Science 2015; 347(6220): 1260419, https://doi.org/10.1126/science.1260419.
- Suba Z. Rosetta stone for cancer cure: comparison of the anticancer capacity of endogenous estrogens, synthetic estrogens and antiestrogens. Oncol Rev 2023; 17: 10708, https://doi.org/10.3389/or.2023.10708.
- Padovan M., Maccari M., Bosio A., De Toni C., Vizzaccaro S., Cestonaro I., Corrà M., Caccese M., Cerretti G., Zagonel V., Lombardi G. Actionable molecular alterations in newly diagnosed and recurrent IDH1/2 wild-type glioblastoma patients and therapeutic implications: a large mono-institutional experience using extensive next-generation sequencing analysis. Eur J Cancer 2023; 191: 112959, https://doi.org/10.1016/j.ejca.2023.112959.
- Huszno J., Grzybowska E. TP53 mutations and SNPs as prognostic and predictive factors in patients with breast cancer. Oncol Lett 2018; 16(1): 34–40, https://doi.org/10.3892/ol.2018.8627.
- Robertson L.B., Armstrong G.N., Olver B.D., Lloyd A.L., Shete S., Lau C., Claus E.B., Barnholtz-Sloan J., Lai R., Il’yasova D., Schildkraut J., Bernstein J.L., Olson S.H., Jenkins R.B., Yang P., Rynearson A.L., Wrensch M., McCoy L., Wienkce J.K., McCarthy B., Davis F., Vick N.A., Johansen C., Bødtcher H., Sadetzki S., Bruchim R.B., Yechezkel G.H., Andersson U., Melin B.S., Bondy M.L., Houlston R.S. Survey of familial glioma and role of germline p16INK4A/p14ARF and p53 mutation. Fam Cancer 2010; 9(3): 413–421, https://doi.org/10.1007/s10689-010-9346-5.
- Maltzman W., Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol 1984; 4(9): 1689–1694, https://doi.org/10.1128/mcb.4.9.1689-1694.1984.
- Park S.K., Park S., Pentek C., Liebman S.W. Tumor suppressor protein p53 expressed in yeast can remain diffuse, form a prion, or form unstable liquid-like droplets. iScience 2020; 24(1): 102000, https://doi.org/10.1016/j.isci.2020.102000.
- Navalkar A., Ghosh S., Pandey S., Paul A., Datta D., Maji S.K. Prion-like p53 amyloids in cancer. Biochemistry 2020; 59(2): 146–155, https://doi.org/10.1021/acs.biochem.9b00796.
- Levine A.J., Puzio-Kuter A.M., Chan C.S., Hainaut P. The role of the p53 protein in stem-cell biology and epigenetic regulation. Cold Spring Harb Perspect Med 2016; 6(9): a026153, https://doi.org/10.1101/cshperspect.a026153.
- Hill K.A., Sommer S.S. p53 as a mutagen test in breast cancer. Environ Mol Mutagen 2002; 39(2–3): 216–227, https://doi.org/10.1002/em.10065.
- Mirzaei S., Paskeh M.D.A., Entezari M., Mirmazloomi S.R., Hassanpoor A., Aboutalebi M., Rezaei S., Hejazi E.S., Kakavand A., Heidari H., Salimimoghadam S., Taheriazam A., Hashemi M., Samarghandian S. SOX2 function in cancers: association with growth, invasion, stemness and therapy response. Biomed Pharmacother 2022; 156: 113860, https://doi.org/10.1016/j.biopha.2022.113860.
- Zhu Y., Huang S., Chen S., Chen J., Wang Z., Wang Y., Zheng H. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis 2021; 12(5): 449, https://doi.org/10.1038/s41419-021-03733-5.
- Zhang S., Xiong X., Sun Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct Target Ther 2020; 5(1): 135, https://doi.org/10.1038/s41392-020-00242-3.
- Abatti L.E., Lado-Fernández P., Huynh L., Collado M., Hoffman M.M., Mitchell J.A. Epigenetic reprogramming of a distal developmental enhancer cluster drives SOX2 overexpression in breast and lung adenocarcinoma. Nucleic Acids Res 2023; 51(19): 10109–10131, https://doi.org/10.1093/nar/gkad734.
- Bao L., Li X., Lin Z. PTEN overexpression promotes glioblastoma death through triggering mitochondrial division and inactivating the Akt pathway. J Recept Signal Transduct Res 2019; 39(3): 215–225, https://doi.org/10.1080/10799893.2019.1655051.
- Yokoi A., Minami M., Hashimura M., Oguri Y., Matsumoto T., Hasegawa Y., Nakagawa M., Ishibashi Y., Ito T., Ohhigata K., Harada Y., Fukagawa N., Saegusa M. PTEN overexpression and nuclear β-catenin stabilization promote morular differentiation through induction of epithelial-mesenchymal transition and cancer stem cell-like properties in endometrial carcinoma. Cell Commun Signal 2022; 20(1): 181, https://doi.org/10.1186/s12964-022-00999-w.
- Li B., Zhang J., Su Y., Hou Y., Wang Z., Zhao L., Sun S., Fu H. Overexpression of PTEN may increase the effect of pemetrexed on A549 cells via inhibition of the PI3K/AKT/mTOR pathway and carbohydrate metabolism. Mol Med Rep 2019; 20(4): 3793–3801, https://doi.org/10.3892/mmr.2019.10617.
- Romagosa C., Simonetti S., López-Vicente L., Mazo A., Lleonart M.E., Castellvi J., Ramon y Cajal S. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 2011; 30(18): 2087–2097, https://doi.org/10.1038/onc.2010.614.
- Yehia L., Keel E., Eng C. The clinical spectrum of PTEN mutations. Annu Rev Med 2020; 71: 103–116, https://doi.org/10.1146/annurev-med-052218-125823.
- Cabrita R., Mitra S., Sanna A., Ekedahl H., Lövgren K., Olsson H., Ingvar C., Isaksson K., Lauss M., Carneiro A., Jönsson G. The role of PTEN loss in immune escape, melanoma prognosis and therapy response. Cancers (Basel) 2020; 12(3): 742, https://doi.org/10.3390/cancers12030742.
- Pentheroudakis G., Mavroeidis L., Papadopoulou K., Koliou G.A., Bamia C., Chatzopoulos K., Samantas E., Mauri D., Efstratiou I., Pectasides D., Makatsoris T., Bafaloukos D., Papakostas P., Papatsibas G., Bombolaki I., Chrisafi S., Kourea H.P., Petraki K., Kafiri G., Fountzilas G., Kotoula V. Angiogenic and antiangiogenic VEGFA splice variants in colorectal cancer: prospective retrospective cohort study in patients treated with irinotecan-based chemotherapy and bevacizumab. Clin Colorectal Cancer 2019; 18(4): e370–e384, https://doi.org/10.1016/j.clcc.2019.07.007.
- 86GDNF glial cell derived neurotrophic factor. URL: https://www.ncbi.nlm.nih.gov/gene/2668.
- Vora P., Venugopal C., Salim S.K., Tatari N., Bakhshinyan D., Singh M., Seyfrid M., Upreti D., Rentas S., Wong N., Williams R., Qazi M.A., Chokshi C., Ding A., Subapanditha M., Savage N., Mahendram S., Ford E., Adile A.A., McKenna D., McFarlane N., Huynh V., Wylie R.G., Pan J., Bramson J., Hope K., Moffat J., Singh S. The rational development of CD133-targeting immunotherapies for glioblastoma. Cell Stem Cell 2020; 26(6): 832–844.e6, https://doi.org/10.1016/j.stem.2020.04.008.
- Irollo E., Pirozzi G. CD133: to be or not to be, is this the real question? Am J Transl Res 2013; 5(6): 563–581.
- Razmara M., Heldin C.H., Lennartsson J. Platelet-derived growth factor-induced Akt phosphorylation requires mTOR/Rictor and phospholipase C-γ1, whereas S6 phosphorylation depends on mTOR/Raptor and phospholipase D. Cell Commun Signal 2013; 11(1): 3, https://doi.org/10.1186/1478-811X-11-3.
- Ostendorp T., Diez J., Heizmann C.W., Fritz G. The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping. Biochim Biophys Acta 2011; 1813(5): 1083–1091, https://doi.org/10.1016/j.bbamcr.2010.10.006.
- Wang H., Mao X., Ye L., Cheng H., Dai X. The role of the S100 protein family in glioma. J Cancer 2022; 13(10): 3022–3030, https://doi.org/10.7150/jca.73365.
- Yin Y., Stephen C.W., Luciani M.G., Fåhraeus R. p53 Stability and activity is regulated by Mdm2-mediated induction of alternative p53 translation products. Nat Cell Biol 2002; 4(6): 462–467, https://doi.org/10.1038/ncb801.
- Marcel V., Perrier S., Aoubala M., Ageorges S., Groves M.J., Diot A., Fernandes K., Tauro S., Bourdon J.C. Δ160p53 is a novel N-terminal p53 isoform encoded by Δ133p53 transcript. FEBS Lett 2010; 584(21): 4463–4468, https://doi.org/10.1016/j.febslet.2010.10.005.
- Pienkowski T., Kowalczyk T., Cysewski D., Kretowski A., Ciborowski M. Glioma and post-translational modifications: a complex relationship. Biochim Biophys Acta Rev Cancer 2023; 1878(6): 189009, https://doi.org/10.1016/j.bbcan.2023.189009.
- Perl K., Ushakov K., Pozniak Y., Yizhar-Barnea O., Bhonker Y., Shivatzki S., Geiger T., Avraham K.B., Shamir R. Reduced changes in protein compared to mRNA levels across non-proliferating tissues. BMC Genomics 2017; 18(1): 305, https://doi.org/10.1186/s12864-017-3683-9.
- Plante I. Dimethylbenz(a)anthracene-induced mammary tumorigenesis in mice. Methods Cell Biol 2021; 163: 21–44, https://doi.org/10.1016/bs.mcb.2020.09.003.
- Avtsyn A.P. Old and new concepts in the teaching on preglioma. Arkh Patol 1972, 34(11): 3–11.
- Kucheryavenko A.S., Chernomyrdin N.V., Gavdush A.A., Alekseeva A.I., Nikitin P.V., Dolganova I.N., Karalkin P.A., Khalansky A.S., Spektor I.E., Skorobogatiy M., Tuchin V.V., Zaytsev K.I. Terahertz dielectric spectroscopy and solid immersion microscopy of ex vivo glioma model 101.8: brain tissue heterogeneity. Biomed Opt Express 2021; 12(8): 5272–5289, https://doi.org/10.1364/BOE.432758.
- Dolganova I.N., Aleksandrova P.V., Nikitin P.V., Alekseeva A.I., Chernomyrdin N.V., Musina G.R., Beshplav S.T., Reshetov I.V., Potapov A.A., Kurlov V.N., Tuchin V.V., Zaytsev K.I. Capability of physically reasonable OCT-based differentiation between intact brain tissues, human brain gliomas of different WHO grades, and glioma model 101.8 from rats. Biomed Opt Express 2020; 11(11): 6780–6798, https://doi.org/10.1364/BOE.409692.
- Kiseleva E.B., Yashin K.S., Moiseev A.A., Timofeeva L.B., Kudelkina V.V., Alekseeva A.I., Meshkova S.V., Polozova A.V., Gelikonov G.V., Zagaynova E.V., Gladkova N.D. Optical coefficients as tools for increasing the optical coherence tomography contrast for normal brain visualization and glioblastoma detection. Neurophotonics 2019; 6(3): 035003, https://doi.org/10.1117/1.NPh.6.3.035003.
- Maksimenko O., Malinovskaya J., Shipulo E., Osipova N., Razzhivina V., Arantseva D., Yarovaya O., Mostovaya U., Khalansky A., Fedoseeva V., Alekseeva A., Vanchugova L., Gorshkova M., Kovalenko E., Balabanyan V., Melnikov P., Baklaushev V., Chekhonin V., Kreuter J., Gelperina S. Doxorubicin-loaded PLGA nanoparticles for the chemotherapy of glioblastoma: towards the pharmaceutical development. Int J Pharm 2019; 572: 118733, https://doi.org/10.1016/j.ijpharm.2019.118733.
- Dzhalilova D.S., Zolotova N.A., Mkhitarov V.A., Kosyreva A.M., Tsvetkov I.S., Khalansky A.S., Alekseeva A.I., Fatkhudinov T.H., Makarova O.V. Morphological and molecular-biological features of glioblastoma progression in tolerant and susceptible to hypoxia Wistar rats. Sci Rep 2023; 13(1): 12694, https://doi.org/10.1038/s41598-023-39914-9.
- Alekseeva A.I., Sentyabreva A.V., Kudelkina V.V., Miroshnichenko E.A., Ikonnikov A.V., Kopantseva E.E., Kosyreva A.M., Fatkhudinov T.K. Rat glioma 101.8 tissue strain: molecular and morphological features. Int J Mol Sci 2025; 26(18): 8992, https://doi.org/10.3390/ijms26188992.










